Optimizing Memory Consumption During Bulk Uploads
Efficient memory management is a constant challenge when you’re dealing with large-scale vector data. In high-volume ingestion scenarios, even seemingly minor configuration choices can significantly impact stability and performance.
Let’s take a look at the best practices and recommendations to help you optimize memory usage during bulk uploads in Qdrant. We’ll cover scenarios with both dense and sparse vectors, helping your deployments remain performant even under high load and avoiding out-of-memory errors.
Indexing for dense vs. sparse vectors
Dense vectors
Qdrant employs an HNSW-based index for fast similarity search on dense vectors. By default, HNSW is built or updated once the number of unindexed vectors in a segment exceeds a set indexing_threshold. Although it delivers excellent query speed, building or updating the HNSW graph can be resource-intensive if it occurs frequently or across many small segments.
Sparse vectors
Sparse vectors use an inverted index. This index is updated at the time of upsertion, meaning you cannot disable or postpone it for sparse vectors. In most cases, its overhead is smaller than that of building an HNSW graph, but you should still be aware that each upsert triggers a sparse index update.
Bulk upload configuration for dense vectors
When performing high-volume vector ingestion, you have two primary options for handling indexing overhead. You should choose one depending on your specific workload and memory constraints:
- Disable HNSW indexing
To reduce memory and CPU pressure during bulk ingestion, you can disable HNSW indexing entirely by setting "m": 0.
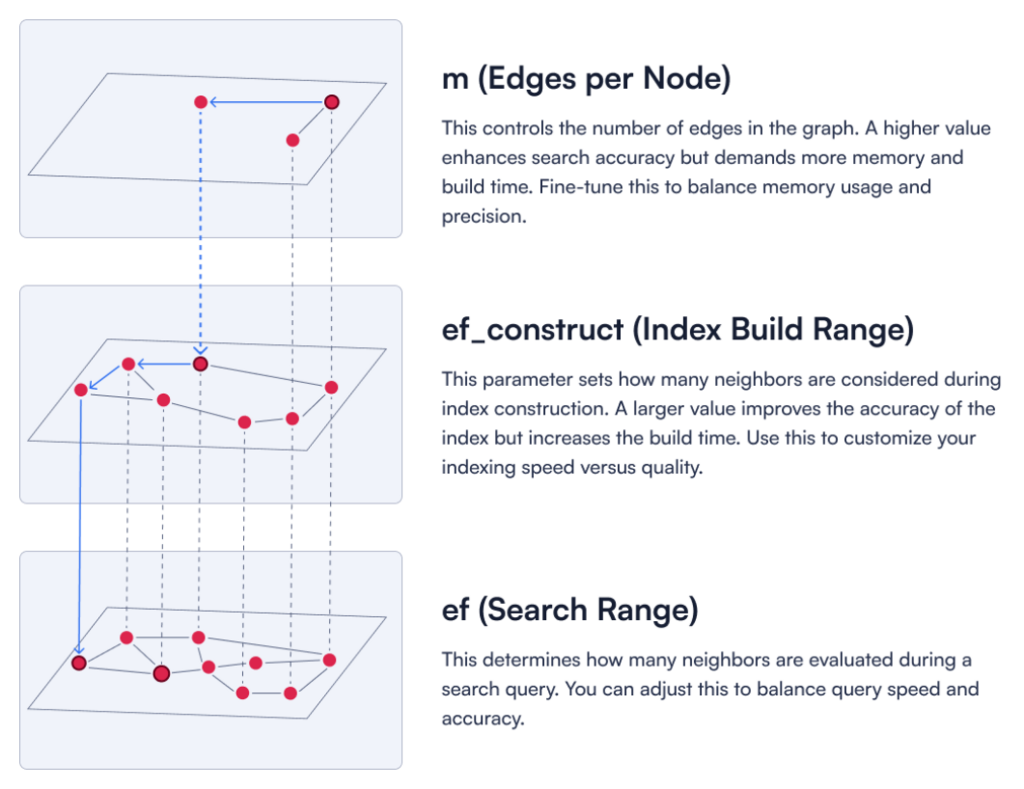
For dense vectors, the m parameter defines how many edges each node in the HNSW graph can have.
This way, no dense vector index will be built, preventing unnecessary CPU usage during ingestion.
Figure 1: A description of three key HNSW parameters.
PATCH /collections/your_collection
{
"hnsw_config": {
"m": 0
}
}
After ingestion is complete, you can re-enable HNSW by setting m back to a production value (commonly 16 or 32).
Remember that search won’t use HNSW until the index is built, so search performance may be slower during this period.
- Disabling optimizations completely
The indexing_threshold tells Qdrant how many unindexed dense vectors can accumulate in a segment before building the HNSW graph. Setting "indexing_threshold"=0 defers indexing entirely, keeping ingestion speed at maximum. However, this means uploaded vectors are not moved to disk while uploading, which can lead to high RAM usage.
PATCH /collections/your_collection
{
"optimizer_config": {
"indexing_threshold": 0
}
}
After bulk ingestion, set indexing_threshold to a positive value to ensure vectors are indexed and searchable via HNSW. Vectors will not be searchable via HNSW until indexing is performed.
Small thresholds (e.g., 100) mean more frequent indexing, which can still be costly if many segments exist. Larger thresholds (e.g., 10000) delay indexing to batch more vectors at once, potentially using more RAM at the moment of index build, but fewer builds overall.
Between these two approaches, we generally recommend disabling HNSW ("m"=0) during bulk ingestion to keep memory usage predictable. Using indexing_threshold=0 can be an alternative, but only if your system has enough memory to accommodate the unindexed vectors in RAM.
On-Disk storage in Qdrant
By default, Qdrant keeps vectors, payload data, and indexes in memory to ensure low-latency queries. However, in large-scale or memory-constrained scenarios, you can configure some or all of them to be stored on-disk. This helps reduce RAM usage at the cost of potential increases in query latency, particularly for cold reads.
When to use on-disk:
- You have very large or rarely used payload data or indexes, and freeing up RAM is worth potential I/O overhead.
- Your dataset doesn’t fit comfortably in available memory.
- You want to reduce memory pressure.
- You can tolerate slower queries if it ensures the system remains stable under heavy loads.
Memmap storage and segmentation
Qdrant uses memory-mapped files (segments) to store data on-disk. Rather than loading all vectors into RAM, Qdrant maps each segment into its address space, paging data in and out on demand. This helps keep the active RAM footprint lower, because data can be paged out if memory pressure is high. But each segment still incurs overhead (metadata, page table entries, etc.).
During high-volume ingestion, you can accumulate dozens of small segments. Qdrant’s optimizer can later merge these into fewer, larger segments, reducing per-segment overhead and lowering total memory usage.
When you create a collection with "on_disk": true, Qdrant will store newly inserted vectors in memmap storage from the start. For example:
PATCH /collections/your_collection
{
"vectors": {
"on_disk": true
}
}
This approach immediately places all incoming vectors on disk, which can be very efficient in case of bulk ingestion.
However, vector data and indexes are stored separately, so enabling on_disk for vectors does not automatically store their indexes on disk. To fully optimize memory usage, you may need to configure both vector storage and index storage independently.
For dense vectors, you can enable on-disk storage for both the vector data and the HNSW index:
PATCH /collections/your_collection
{
"vectors": {
"on_disk": true
},
"hnsw_config": {
"on_disk": true
}
}
For sparse vectors, you need to enable on_disk for both the vector data and the sparse index separately:
PATCH /collections/your_collection
{
"sparse_vectors": {
"text": {
"on_disk": true,
"index": {
"on_disk": true
}
}
}
}
Best practices for high-volume vector ingestion
Bulk ingestion can lead to high memory consumption and even out-of-memory (OOM) errors. If you’re experiencing out-of-memory errors with your current setup, scaling up temporarily (increasing available RAM) will provide a buffer while you adjust Qdrant’s configuration for more a efficient data ingestion.
The key here is to control indexing overhead. Let’s walk through the best practices for high-volume vector ingestion in a constrained-memory environment.
1. Store vector data on disk immediately
The most effective way to reduce memory usage is to store vector data on disk right from the start using on_disk: true. This prevents RAM from being overloaded with raw vectors before optimization kicks in.
PATCH /collections/your_collection
{
"vectors": {
"on_disk": true
}
}
Previously, vector data had to be held in RAM until optimizers could move it to disk, which caused significant memory pressure. Now, by writing vectors to disk directly, memory overhead is significantly reduced, making bulk ingestion much more efficient.
2. Disable HNSW for dense vectors (m=0)
During an initial bulk load, you can disable dense indexing by setting "m": 0. This ensures Qdrant won’t build an HNSW graph for incoming vectors, avoiding unnecessary memory and CPU usage.
PATCH /collections/your_collection
{
"hnsw_config": {
"m": 0
},
"optimizer_config": {
"indexing_threshold": 10000
}
}
3. Let the optimizer run after bulk uploads
Qdrant’s optimizers continuously restructure data to improve search efficiency. However, during a bulk upload, this can lead to excessive data movement and overhead as segments are constantly reorganized while new data is still arriving.
To avoid this, upload all data first, then allow the optimizer to process everything in one go. This minimizes redundant operations and ensures a more efficient segment structure.
4. Wait for indexation to clear up memory
Before performing additional operations, allow Qdrant to finish any ongoing indexing. Large indexing jobs can keep memory usage high until they fully complete.
Monitor Qdrant logs or metrics to confirm when indexing finishes—once that happens, memory consumption should drop as intermediate data structures are freed.
5. Re-enable HNSW post-ingestion
After the ingestion phase is over and memory usage has stabilized, re-enable HNSW for dense vectors by setting m back to a production value (commonly 16 or 32):
PATCH /collections/your_collection
{
"hnsw_config": {
"m": 16
}
}
5. Enable quantization
If you had planned to store all dense vectors on disk, be aware that searches can slow down drastically due to frequent disk I/O while memory pressure is high. A more balanced approach is scalar quantization: compress vectors (e.g., to int8) so they fit in RAM without occupying as much space as full floating-point values.
PATCH /collections/your_collection
{
"quantization_config": {
"scalar": {
"type": "int8",
"always_ram": true
}
}
}
Quantized vectors remain in-memory yet consume less space, preserving much of the performance advantage of RAM-based search. Learn more about vector quantization.
Conclusion
High-volume vector ingestion can place significant memory demands on Qdrant, especially if dense vectors are indexed in real time. By following these tips, you can substantially reduce the risk of out-of-memory errors and maintain stable performance in a memory-limited environment.
As always, monitor your system’s behavior. Review logs, watch metrics, and keep an eye on memory usage. Each workload is different, so it’s wise to fine-tune Qdrant’s parameters according to your hardware and data scale.



