Question Answering as a Service with Cohere and Qdrant
Kacper Łukawski
·November 29, 2022

Bi-encoders are probably the most efficient way of setting up a semantic Question Answering system. This architecture relies on the same neural model that creates vector embeddings for both questions and answers. The assumption is, both question and answer should have representations close to each other in the latent space. It should be like that because they should both describe the same semantic concept. That doesn’t apply to answers like “Yes” or “No” though, but standard FAQ-like problems are a bit easier as there is typically an overlap between both texts. Not necessarily in terms of wording, but in their semantics.
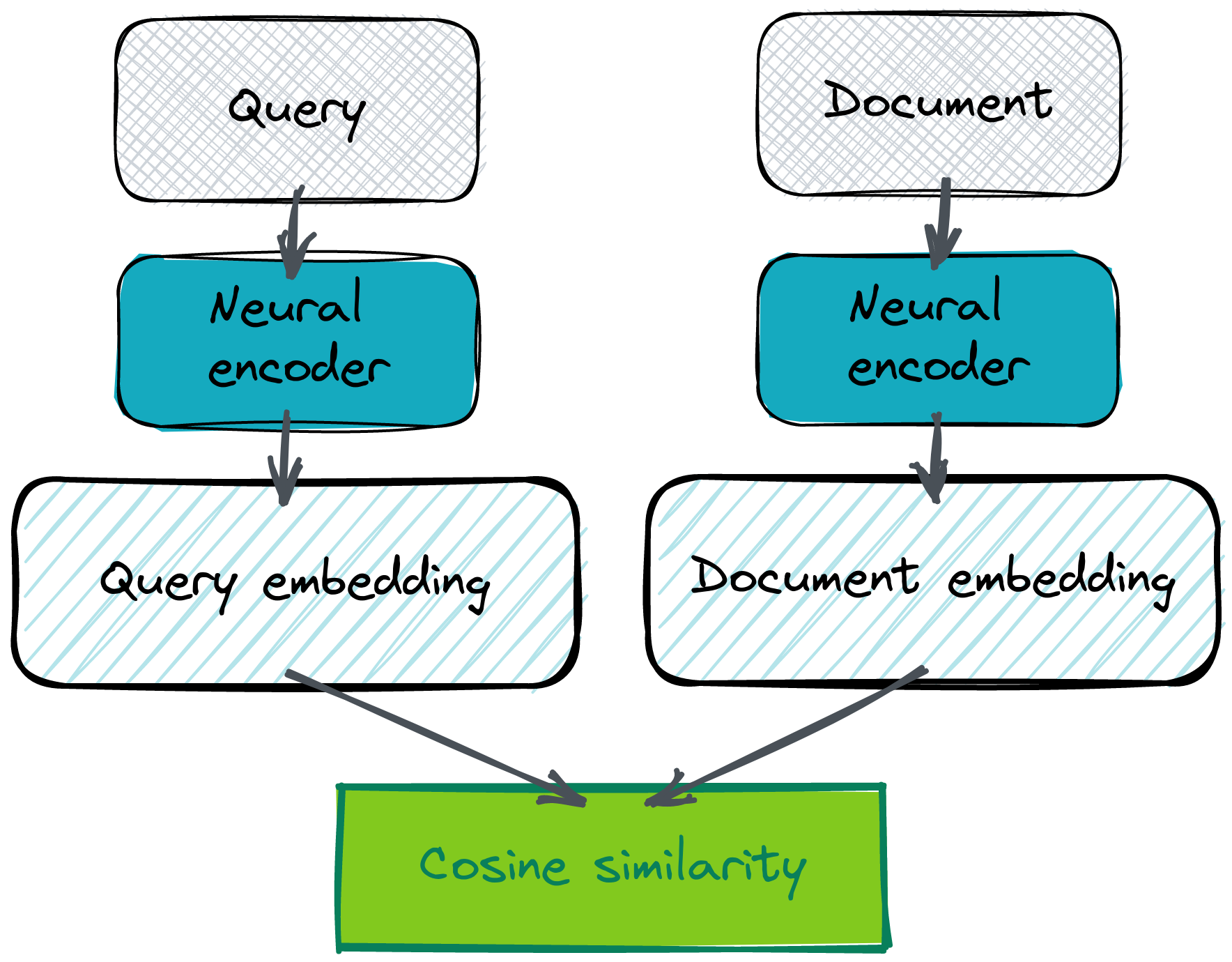
And yeah, you need to bring your own embeddings, in order to even start. There are various ways how to obtain them, but using Cohere co.embed API is probably the easiest and most convenient method.
Why co.embed API and Qdrant go well together?
Maintaining a Large Language Model might be hard and expensive. Scaling it up and down, when the traffic changes, require even more effort and becomes unpredictable. That might be definitely a blocker for any semantic search system. But if you want to start right away, you may consider using a SaaS model, Cohere’s co.embed API in particular. It gives you state-of-the-art language models available as a Highly Available HTTP service with no need to train or maintain your own service. As all the communication is done with JSONs, you can simply provide the co.embed output as Qdrant input.
# Putting the co.embed API response directly as Qdrant method input
qdrant_client.upsert(
collection_name="collection",
points=rest.Batch(
ids=[...],
vectors=cohere_client.embed(...).embeddings,
payloads=[...],
),
)
Both tools are easy to combine, so you can start working with semantic search in a few minutes, not days.
And what if your needs are so specific that you need to fine-tune a general usage model? Co.embed API goes beyond pre-trained encoders and allows providing some custom datasets to customize the embedding model with your own data. As a result, you get the quality of domain-specific models, but without worrying about infrastructure.
System architecture overview
In real systems, answers get vectorized and stored in an efficient vector search database. We typically don’t even need to provide specific answers, but just use sentences or paragraphs of text and vectorize them instead. Still, if a bit longer piece of text contains the answer to a particular question, its distance to the question embedding should not be that far away. And for sure closer than all the other, non-matching answers. Storing the answer embeddings in a vector database makes the search process way easier.
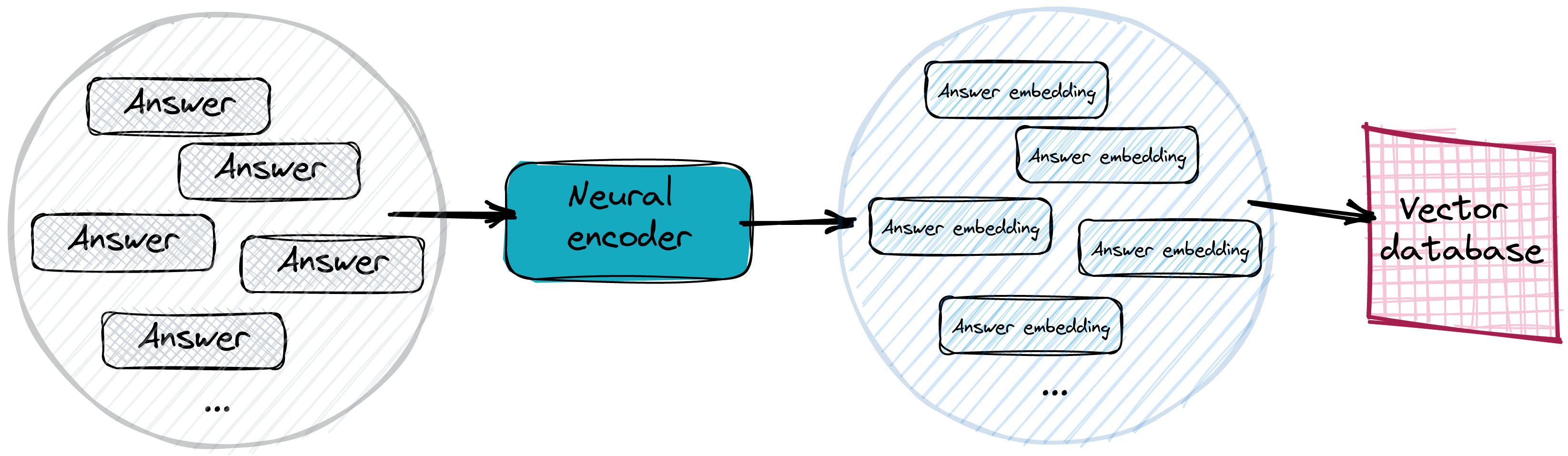
Looking for the correct answer
Once our database is working and all the answer embeddings are already in place, we can start querying it. We basically perform the same vectorization on a given question and ask the database to provide some near neighbours. We rely on the embeddings to be close to each other, so we expect the points with the smallest distance in the latent space to contain the proper answer.
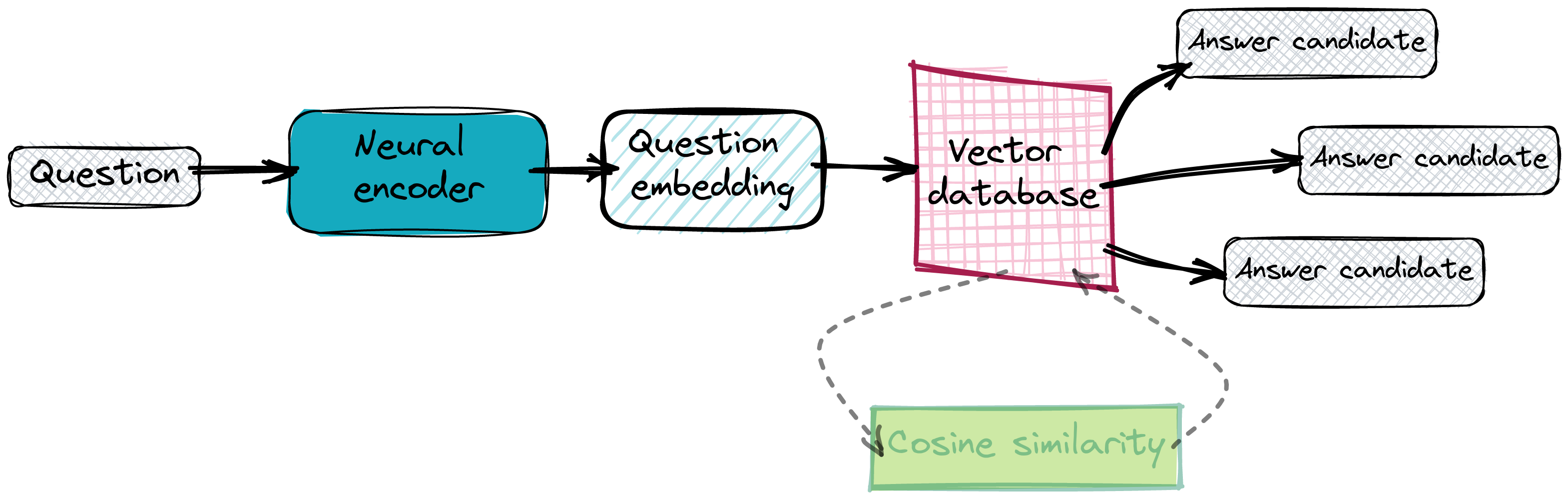
Implementing the QA search system with SaaS tools
We don’t want to maintain our own service for the neural encoder, nor even set up a Qdrant instance. There are SaaS solutions for both — Cohere’s co.embed API and Qdrant Cloud, so we’ll use them instead of on-premise tools.
Question Answering on biomedical data
We’re going to implement the Question Answering system for the biomedical data. There is a pubmed_qa dataset, with it pqa_labeled subset containing 1,000 examples of questions and answers labelled by domain experts. Our system is going to be fed with the embeddings generated by co.embed API and we’ll load them to Qdrant. Using Qdrant Cloud vs your own instance does not matter much here. There is a subtle difference in how to connect to the cloud instance, but all the other operations are executed in the same way.
from datasets import load_dataset
# Loading the dataset from HuggingFace hub. It consists of several columns: pubid,
# question, context, long_answer and final_decision. For the purposes of our system,
# we’ll use question and long_answer.
dataset = load_dataset("pubmed_qa", "pqa_labeled")
| pubid | question | context | long_answer | final_decision |
|---|---|---|---|---|
| 18802997 | Can calprotectin predict relapse risk in infla… | … | Measuring calprotectin may help to identify UC… | maybe |
| 20538207 | Should temperature be monitorized during kidne… | … | The new storage can affords more stable temper… | no |
| 25521278 | Is plate clearing a risk factor for obesity? | … | The tendency to clear one’s plate when eating … | yes |
| 17595200 | Is there an intrauterine influence on obesity? | … | Comparison of mother-offspring and father-offs.. | no |
| 15280782 | Is unsafe sexual behaviour increasing among HI… | … | There was no evidence of a trend in unsafe sex… | no |
Using Cohere and Qdrant to build the answers database
In order to start generating the embeddings, you need to create a Cohere account. That will start your trial period, so you’ll be able to vectorize the texts for free. Once logged in, your default API key will be available in Settings. We’ll need it to call the co.embed API. with the official python package.
import cohere
cohere_client = cohere.Client(COHERE_API_KEY)
# Generating the embeddings with Cohere client library
embeddings = cohere_client.embed(
texts=["A test sentence"],
model="large",
)
vector_size = len(embeddings.embeddings[0])
print(vector_size) # output: 4096
Let’s connect to the Qdrant instance first and create a collection with the proper configuration, so we can put some embeddings into it later on.
# Connecting to Qdrant Cloud with qdrant-client requires providing the api_key.
# If you use an on-premise instance, it has to be skipped.
qdrant_client = QdrantClient(
host="xyz-example.eu-central.aws.cloud.qdrant.io",
prefer_grpc=True,
api_key=QDRANT_API_KEY,
)
Now we’re able to vectorize all the answers. They are going to form our collection, so we can also put them already into Qdrant, along with the payloads and identifiers. That will make our dataset easily searchable.
answer_response = cohere_client.embed(
texts=dataset["train"]["long_answer"],
model="large",
)
vectors = [
# Conversion to float is required for Qdrant
list(map(float, vector))
for vector in answer_response.embeddings
]
ids = [entry["pubid"] for entry in dataset["train"]]
# Filling up Qdrant collection with the embeddings generated by Cohere co.embed API
qdrant_client.upsert(
collection_name="pubmed_qa",
points=rest.Batch(
ids=ids,
vectors=vectors,
payloads=list(dataset["train"]),
)
)
And that’s it. Without even setting up a single server on our own, we created a system that might be easily asked a question. I don’t want to call it serverless, as this term is already taken, but co.embed API with Qdrant Cloud makes everything way easier to maintain.
Answering the questions with semantic search — the quality
It’s high time to query our database with some questions. It might be interesting to somehow measure the quality of the system in general. In those kinds of problems we typically use top-k accuracy. We assume the prediction of the system was correct if the correct answer was present in the first k results.
# Finding the position at which Qdrant provided the expected answer for each question.
# That allows to calculate accuracy@k for different values of k.
k_max = 10
answer_positions = []
for embedding, pubid in tqdm(zip(question_response.embeddings, ids)):
response = qdrant_client.search(
collection_name="pubmed_qa",
query_vector=embedding,
limit=k_max,
)
answer_ids = [record.id for record in response]
if pubid in answer_ids:
answer_positions.append(answer_ids.index(pubid))
else:
answer_positions.append(-1)
Saved answer positions allow us to calculate the metric for different k values.
# Prepared answer positions are being used to calculate different values of accuracy@k
for k in range(1, k_max + 1):
correct_answers = len(
list(
filter(lambda x: 0 <= x < k, answer_positions)
)
)
print(f"accuracy@{k} =", correct_answers / len(dataset["train"]))
Here are the values of the top-k accuracy for different values of k:
| metric | value |
|---|---|
| accuracy@1 | 0.877 |
| accuracy@2 | 0.921 |
| accuracy@3 | 0.942 |
| accuracy@4 | 0.950 |
| accuracy@5 | 0.956 |
| accuracy@6 | 0.960 |
| accuracy@7 | 0.964 |
| accuracy@8 | 0.971 |
| accuracy@9 | 0.976 |
| accuracy@10 | 0.977 |
It seems like our system worked pretty well even if we consider just the first result, with the lowest distance. We failed with around 12% of questions. But numbers become better with the higher values of k. It might be also valuable to check out what questions our system failed to answer, their perfect match and our guesses.
We managed to implement a working Question Answering system within just a few lines of code. If you are fine with the results achieved, then you can start using it right away. Still, if you feel you need a slight improvement, then fine-tuning the model is a way to go. If you want to check out the full source code, it is available on Google Colab.
