Building a Chain-of-Thought Medical Chatbot with Qdrant and DSPy
Accessing medical information from LLMs can lead to hallucinations or outdated information. Relying on this type of information can result in serious medical consequences. Building a trustworthy and context-aware medical chatbot can solve this.
In this article, we will look at how to tackle these challenges using:
- Retrieval-Augmented Generation (RAG): Instead of answering the questions from scratch, the bot retrieves the information from medical literature before answering questions.
- Filtering: Users can filter the results by specialty and publication year, ensuring the information is accurate and up-to-date.
Let’s discover the technologies needed to build the medical bot.
Tech Stack Overview
To build a robust and trustworthy medical chatbot, we will combine the following technologies:
- Qdrant Cloud: Qdrant is a high-performance vector search engine for storing and retrieving large collections of embeddings. In this project, we will use it to enable fast and accurate search across millions of medical documents, supporting dense and multi-vector (ColBERT) retrieval for context-aware answers.
- Stanford DSPy: DSPy is the AI framework we will use to obtain the final answer. It allows the medical bot to retrieve the relevant information and reason step-by-step to produce accurate and explainable answers.
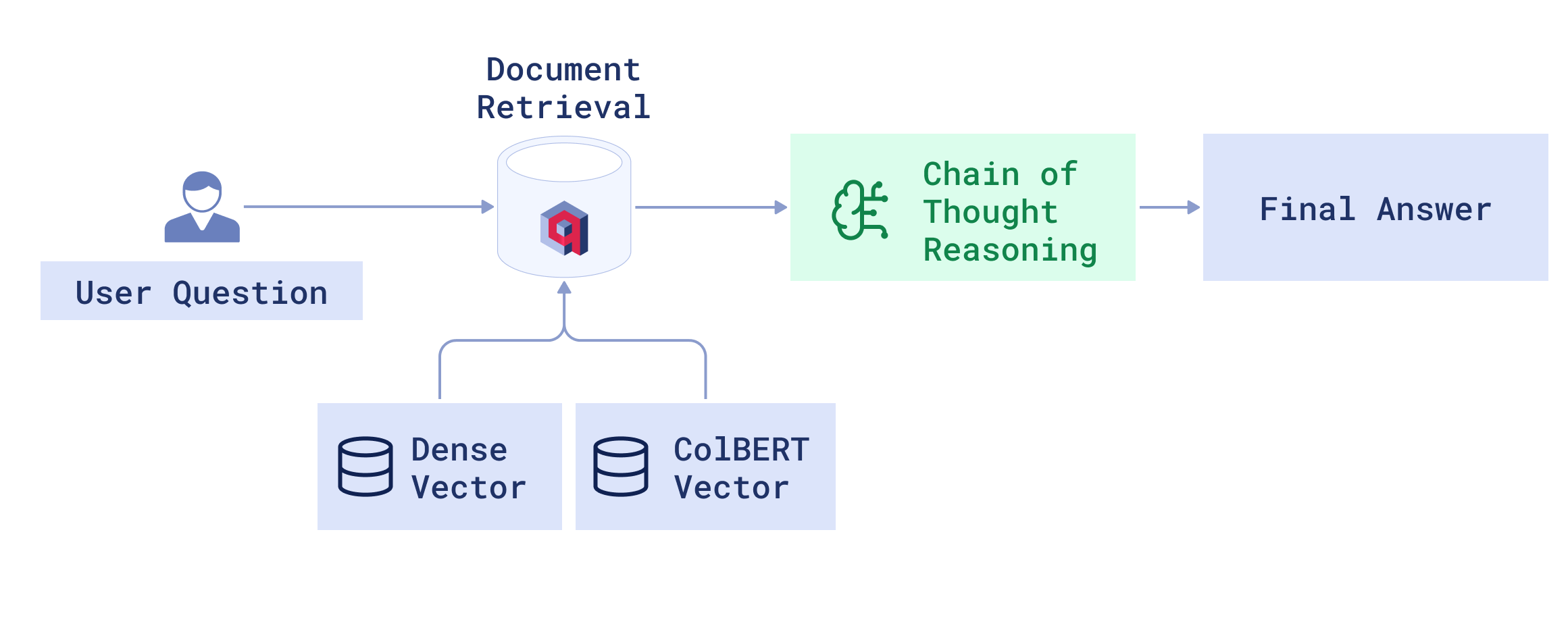
Dataset Preparation and Indexing
A medical chatbot is only as good as the knowledge it has access to. For this project, we will leverage the MIRIAD medical dataset, a large-scale collection of medical passages enriched with metadata such as publication year and specialty.
Indexing with Dense and ColBERT Multivectors
To enable high-quality retrieval, we will embed each medical passage with two models:
- Dense Embeddings: These are generated using the
BAAI/bge-small-enmodel and capture the passages’ general semantic meaning. - ColBERT Multivectors: These provide more fine-grained representations, enabling precise ranking of results.
dense_documents = [
models.Document(text=doc, model="BAAI/bge-small-en") for doc in ds["passage_text"]
]
colbert_documents = [
models.Document(text=doc, model="colbert-ir/colbertv2.0")
for doc in ds["passage_text"]
]
collection_name = "miriad"
# Create collection
if not client.collection_exists(collection_name):
client.create_collection(
collection_name=collection_name,
vectors_config={
"dense": models.VectorParams(size=384, distance=models.Distance.COSINE),
"colbert": models.VectorParams(
size=128,
distance=models.Distance.COSINE,
multivector_config=models.MultiVectorConfig(
comparator=models.MultiVectorComparator.MAX_SIM
),
hnsw_config=models.HnswConfigDiff(m=0), # reranker: no indexing
),
},
)
We disable indexing for the ColBERT multivector since it will only be used for reranking. To learn more about this, check out the How to Effectively Use Multivector Representations in Qdrant for Reranking article.
Batch Uploading to Qdrant
To avoid hitting API limits, we upload the data in batches, each batch containing:
- The passage text
- ColBERT and dense embeddings.
yearandspecialtymetadata fields.
BATCH_SIZE = 3
points_batch = []
for i in range(len(ds["passage_text"])):
point = models.PointStruct(
id=i,
vector={"dense": dense_documents[i], "colbert": colbert_documents[i]},
payload={
"passage_text": ds["passage_text"][i],
"year": ds["year"][i],
"specialty": ds["specialty"][i],
},
)
points_batch.append(point)
if len(points_batch) == BATCH_SIZE:
client.upsert(collection_name=collection_name, points=points_batch)
print(f"Uploaded batch ending at index {i}")
points_batch = []
# Final flush
if points_batch:
client.upsert(collection_name=collection_name, points=points_batch)
print("Uploaded final batch.")
Retrieval-Augmented Generation (RAG) Pipeline
Our chatbot will use a Retrieval-Augmented Generation (RAG) pipeline to ensure its answers are grounded in medical literature.
Integration of DSPy and Qdrant
At the heart of the application is the Qdrant vector database that provides the information sent to DSPy to generate the final answer. This is what happens when a user submits a query:
- DSPy searches against the Qdrant vector database to retrieve the top documents and answers the query. The results are also filtered with a particular year range for a specific specialty.
- The retrieved passages are then reranked using ColBERT multivector embeddings, leading to the most relevant and contextually appropriate answers.
- DSPy uses these passages to guide the language model through a chain-of-thought reasoning to generate the most accurate answer.
def rerank_with_colbert(query_text, min_year, max_year, specialty):
from fastembed import TextEmbedding, LateInteractionTextEmbedding
# Encode query once with both models
dense_model = TextEmbedding("BAAI/bge-small-en")
colbert_model = LateInteractionTextEmbedding("colbert-ir/colbertv2.0")
dense_query = list(dense_model.embed(query_text))[0]
colbert_query = list(colbert_model.embed(query_text))[0]
# Combined query: retrieve with dense,
# rerank with ColBERT
results = client.query_points(
collection_name=collection_name,
prefetch=models.Prefetch(query=dense_query, using="dense"),
query=colbert_query,
using="colbert",
limit=5,
with_payload=True,
query_filter=Filter(
must=[
FieldCondition(key="specialty", match=MatchValue(value=specialty)),
FieldCondition(
key="year",
range=models.Range(gt=None, gte=min_year, lt=None, lte=max_year),
),
]
),
)
points = results.points
docs = []
for point in points:
docs.append(point.payload["passage_text"])
return docs
The pipeline ensures that each response is grounded in real and recent medical literature and is aligned with the user’s needs.
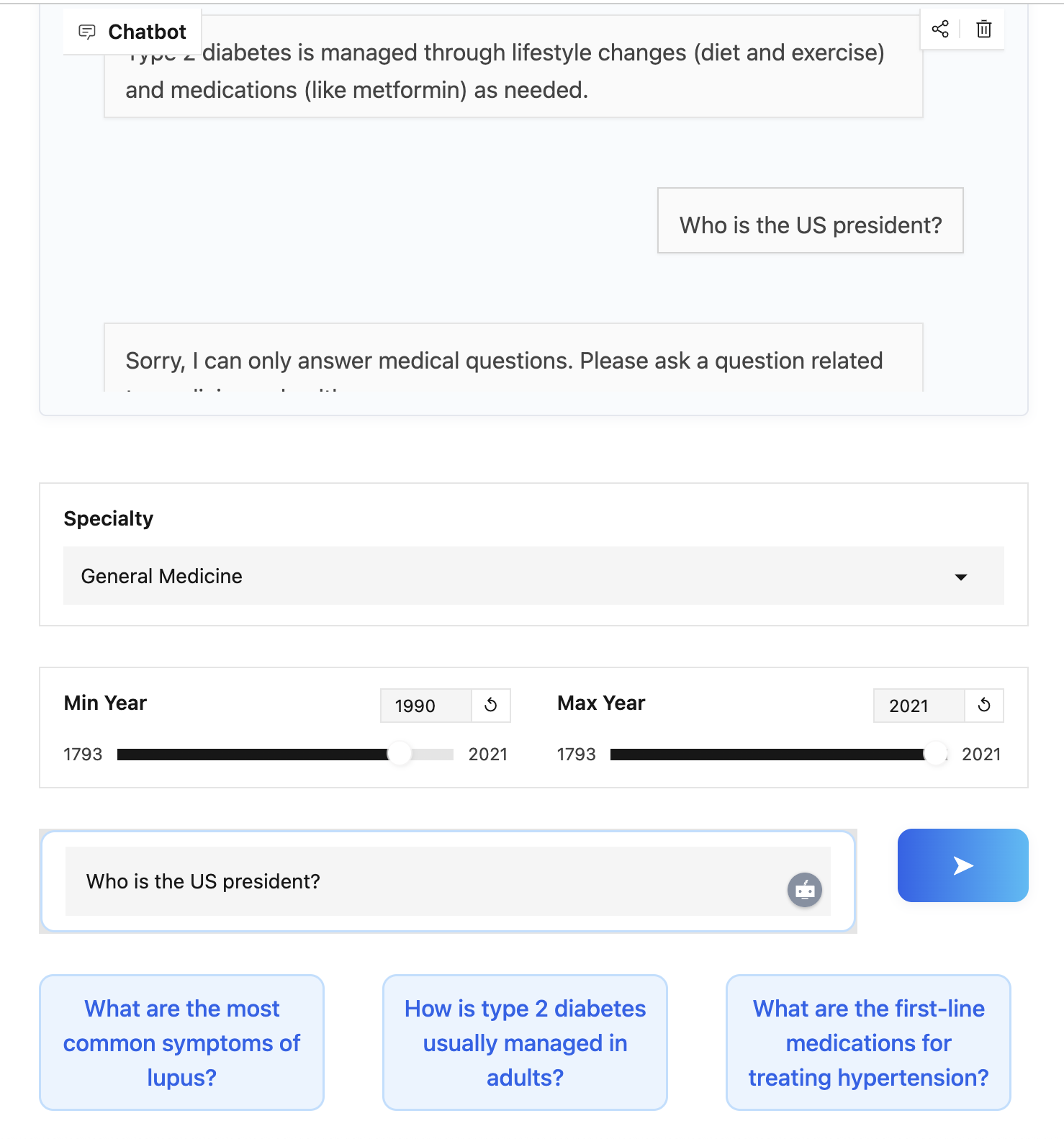
Guardrails and Medical Question Detection
Since this is a medical chatbot, we can introduce a simple guardrail to ensure it doesn’t respond to unrelated questions like the weather. This can be implemented using a DSPy module.
The chatbot checks if every question is medical-related before attempting to answer it. This is achieved by a DSPy module that classifies each incoming query as medical or not. If the question is not medical-related, the chatbot declines to answer, reducing the risk of misinformation or inappropriate responses.
class MedicalGuardrail(dspy.Module):
def forward(self, question):
prompt = (
"""
Is the following question a medical question?
Answer with 'Yes' or 'No'.n"
f"Question: {question}n"
"Answer:
"""
)
response = dspy.settings.lm(prompt)
answer = response[0].strip().lower()
return answer.startswith("yes")
if not self.guardrail.forward(question):
class DummyResult:
final_answer = """
Sorry, I can only answer medical questions.
Please ask a question related to medicine or healthcare
"""
return DummyResult()
By combining this guardrail with specialty and year filtering, we ensure that the chatbot:
- Only answers medical questions.
- Answers questions from recent medical literature.
- Doesn’t make up answers by grounding its answers in the provided literature.
Conclusion
By leveraging Qdrant and DSPy, you can build a medical chatbot that generates accurate and up-to-date medical responses. Qdrant provides the technology and enables fast and scalable retrieval, while DSPy synthesizes this information to provide correct answers grounded in the medical literature. As a result, you can achieve a medical system that is truthful, safe, and provides relevant responses. Check out the entire project from this notebook. You’ll need a free Qdrant Cloud account to run the notebook.


